Usage guide:
1. User input should include two parts, and the output will have two content.
Input 1: Select a reference arrangement (a benchmark arrangement) for mitochondrial genome organization.
(1) In this step, our web server provides two reference genomes, typical vertebrate arrangement and typical invertebrate arrangement, respectively (the first option);
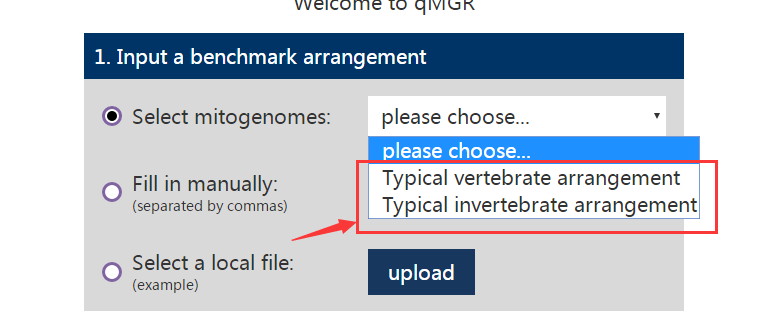
(2) The second option is that the user also manually enters the reference gene order by himself. And the user can also modify it by copying the order of the genes in the example.
(3) User can also create a local reference arrangement file according to the example we provide (click on the example to download it), and directly upload the local file to complete the input request of this step.
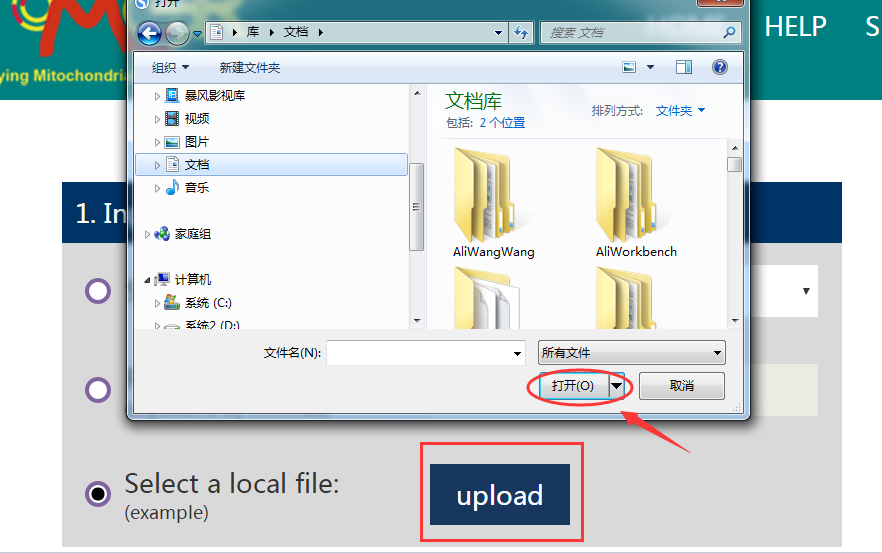
Note: When we study the mitochondrial genome rearrangement in vertebrates, we can choose the typical vertebrate arrangement as the benchmark. Correspondingly, we will select invertebrates as reference arrangement when studying invertebrates. And sometimes, we can also choose other arrangements as long as they are the highest frequency arrangement types in a certain group.
2. Input 2: Enter one or more types of mitogenome arrangement to be studied.
(1) User can enter the gene order of different mitogenomes (or different species) by species classification. In this case, a single time is often the frequency of rearrangement type of each species or mitogenome;
(2) In this section, if the user enters each entry according to the type of arrangement, then the same types may exist more than once, merge them, and add the frequency of occurrence to the front of each actual genetic arrangement type. The way helps our server output the results faster.
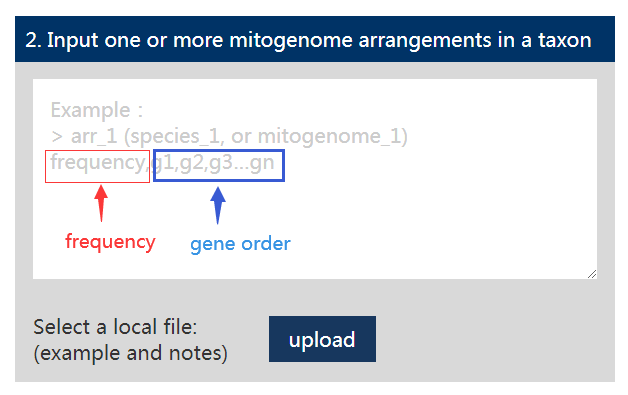
Our server also gives a template and documentation for the input file 2(example and notes), user can refer to this file for more information on the preparation of the file.
①: Arrangement frequency; ② Gene order; ③: Description

3. Output:
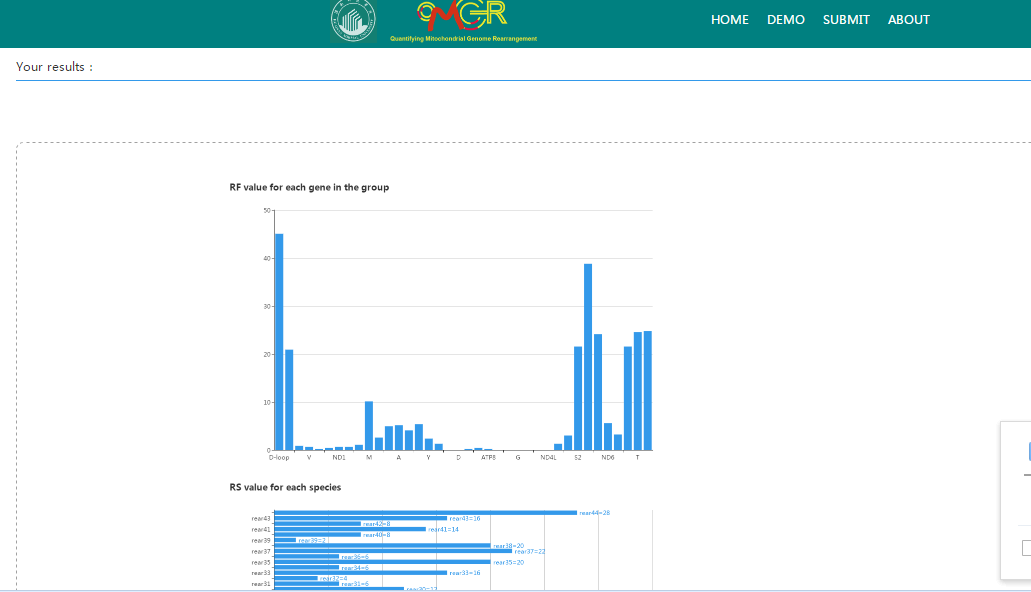
The output includes: (1) the rearrangement frequency (RF)of each individual gene in the taxonomic group, and (2) the rearrangement score (RS) of each output species or rearrangement type. They are presented in graphical and numerical form, respectively. And the results can also be downloaded directly.
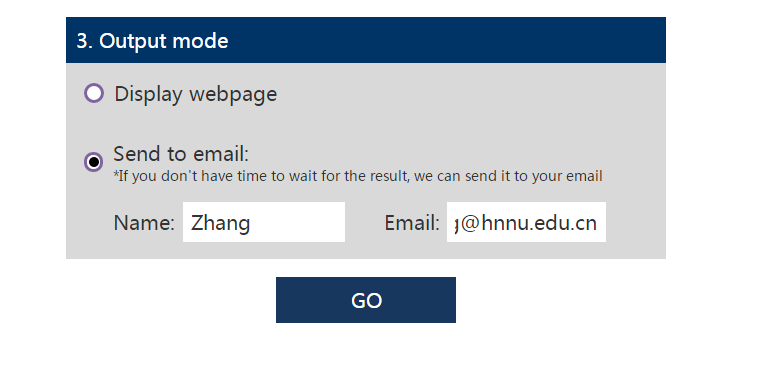
In order to facilitate the user, the presentation form of the result may also have two alternatives, one is to directly display on the webpage, and the other is to send to the user's mailbox.
* The first version of our web server, the home page and the submit page, can implement the qMGR algorithm.



