1. About qMGR
Welcome to qMGR. It is a free web server for quantifying mitochondrial genome rearrangement. The qMGR can calculate the rearrangement frequency (RF) of individual genes within a certain group relative to the benchmark, as well as the total rearrangement score (RS) of a species or a mitogenome. See Figure 1 and our publication for more details on the method.
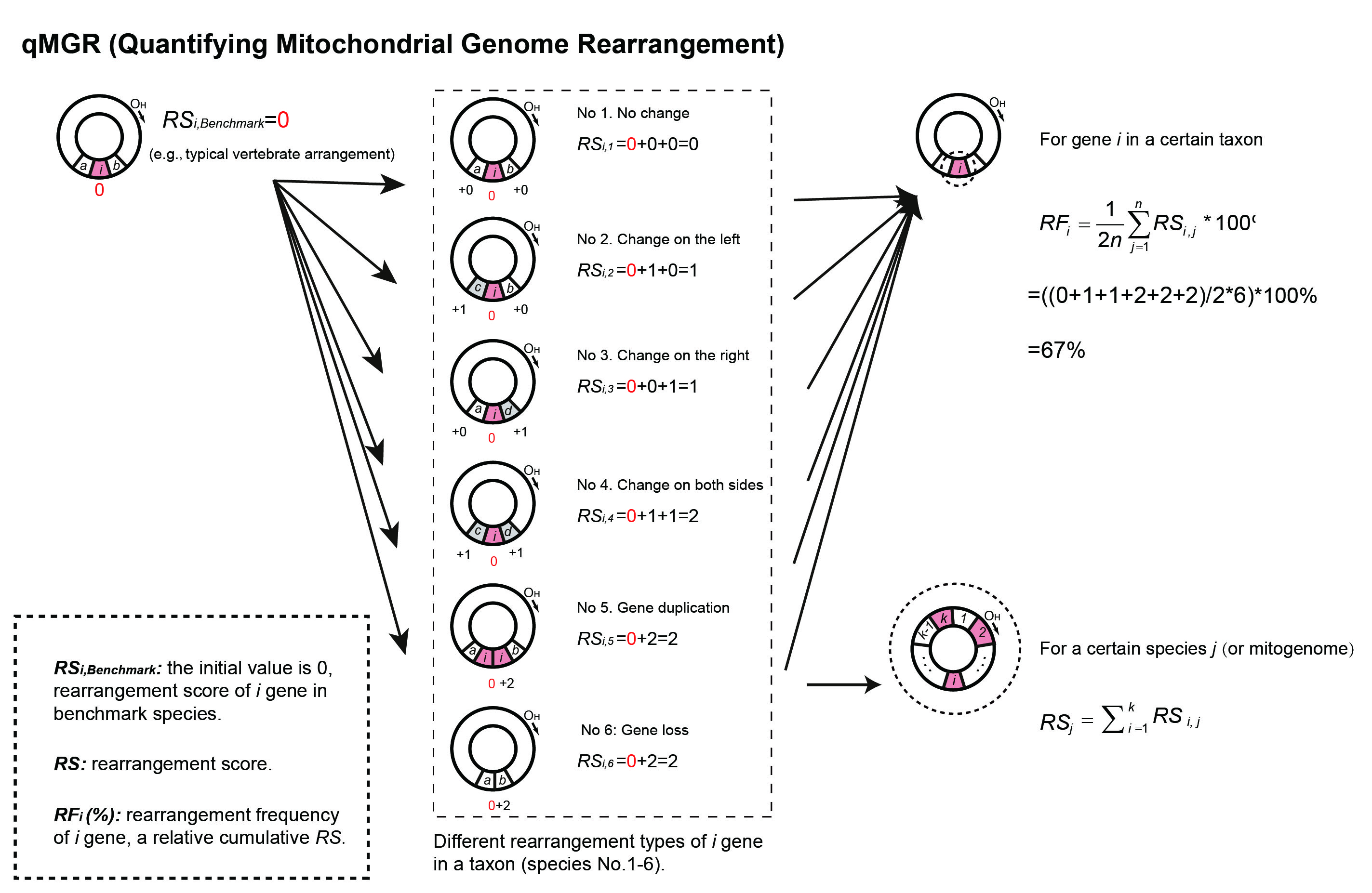
Figure 1. Schematic diagram of qMGR to calculate RS and RF of a given gene i in a given taxon.
3. FAQ
Q: What is mitochondrial genome rearrangement?
A: Compared to a reference
arrangement, the baseline rearrangement, the researchers usually examine whether the gene order on the mitogenomes of the
species we are interested in has changed, including gene transposition, gene
inversion, gene duplication, and gene loss.
Q: How does qMGR calculate mitochondrial genome rearrangements?
A: we proposed a novel statistical method called “Accumulated Neighbor Changes” to calculate relative cumulative scores for each rearranged gene. As long as the left and right neighbors of a given gene change, we will give it 1-2 points, see our publication for more details.
Q: What aspects of genome rearrangement can be detected by qMGR?
A: It can calculate a rearrangement frequency (RF) of a single gene in a certain animal group, as well as the total rearrangement score (RS) of a species or a mitogenome.
Q: Can qMGR examine the rearrangement of a certain type of gene in the mitochondrial genome, such as protein-coding genes, tRNA genes?
A: Yes, as long as the number of genes to be investigated is consistent with their number of reference mitogenome.
Q: Can qMGR examine the rearrangement of pseudogenes and the origin of L-strand replication (OL) ?
A: Yes, it does. However, we do not recommend monitoring the rearrangement of a pseudogene because the presence of pseudogenes is not universal, although pseudogenes may also increase the rearrangement score (RS) of their corresponding genes. For the OL, when you consider its relative position in the reference arrangement and the mitogenome arrangements of a certain group to be studied, the rearrangement of the OL is achievable.
Q: What can I do if qMGR produced an error or is behaving unexpectedly?
A: It would be nice if you inform us by sending an email to jifengzhang {at} hnnu.edu.cn.
4. Quick links
Organelle Genome Resources of NCBI (https://www.ncbi.nlm.nih.gov/genome/organelle/)
MITOS (http://mitos.bioinf.uni-leipzig.de/)
tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/)
mitotRNAdb (http://mttrna.bioinf.uni-leipzig.de/mtDataOutput/Welcome)
5. Bug Report or Question?
If you have spot an error in any of our web server, have a question or suggestions for us, we would be delighted if you would please contact us at jifengzhang {at} hnnu.edu.cn.
6. Citation:
Jifeng Zhang, et al. qMGR: a new approach for quantifying mitochondrial genome rearrangement[J]. Mitochondrion, 2020. (https://doi.org/10.1016/j.mito.2020.02.004)


